Chapter 6 Basic Regression
Now that we are equipped with data visualization skills from Chapter 3, data wrangling skills from Chapter 4, and an understanding of “tidy” data format from Chapter 5, let’s now proceed with data modeling. The fundamental premise of data modeling is to make explicit the relationship between:
- an outcome variable \(y\), also called a dependent variable or response variable and
- an explanatory/predictor variable \(x\), also called an independent variable or covariate.
Another way to state this is using mathematical terminology: we will model the outcome variable \(y\) “as a function” of the explanatory/predictor variable \(x\). When we say “function” here, we aren’t referring to a function like the ggplot() function, but rather as a mathematical function. Why do we have two different labels, explanatory and predictor, for the variable \(x\)? That’s because even though the two terms are used interchangeably, roughly speaking data modeling serves one of two purposes:
- Modeling for explanation: When you want to explicitly describe and quantify the relationship between an outcome variable \(y\) and a set of explanatory variables \(x\), determine the significance of any relationships, and have measures summarizing these relationships.
- Modeling for prediction: When you want to predict an outcome variable \(y\) based on the information contained in a set of predictor variables. Unlike modeling for explanation however, you don’t care so much about understanding how all the variables relate and interact with one another, but rather only whether you can make good predictions about \(y\) using the information in predictor variables \(x\).
For example, say you are interested in an outcome variable \(y\) of whether patients develop lung cancer and information \(x\) on their risk factors, such as smoking habits, age, and socioeconomic status. If we are modeling for explanation, we would be interested describing and quantifying the effects of the different risk factors. One reason could be because you want to design an intervention to reduce lung cancer incidence in a population, such as targeting smokers of a specific age group with advertising for smoking cessation programs. If we are modeling for prediction however, we wouldn’t care so much about understanding how all the individual risk factors contribute to lung cancer incidence, but rather only whether we can make good predictions of who will contract lung cancer.
In this book, we’ll focus on modeling for explanation and hence refer to \(x\). If you are interested in learning about modeling for prediction, we suggest you check out books and courses on machine learning. Furthermore, while there exists many techniques for modeling, such as tree-based models and neural networks, in this book we’ll focus on one particular technique: linear regression, one of the most commonly-used and easy-to-understand approaches to modeling.
Linear regression involves a numerical outcome variable \(y\) and explanatory variables \(x\) that are either numerical or categorical. Furthermore, the relationship between \(y\) and \(x\) is assumed to be linear, or in other words, a line. However we’ll see that what constitutes a “line” will vary depending on the nature of your \(x\) explanatory variables. We’ll study
- In Chapter 6 on basic regression, we’ll only consider models with a single explanatory variable \(x\)
- In Chapter 7 on multiple regression, we’ll consider models with two explanatory variables \(x_1\) and \(x_2\):
- In Chapter 11 on inference for regression, we’ll revisit our regression models and analyze the results using the tools for “statistical inference” you’ll develop in Chapters 8, 9, and 10 on sampling, confidence intervals, and hypothesis test/p-values respectively.
Let’s now begin with basic regression, which are linear regression models with a single explanatory variable \(x\). We’ll also discuss important statistical concepts like the correlation coefficient, that “correlation isn’t necessarily causation”, and what it means for a line to be “best fitting.”
Needed packages
Let’s now load all the packages needed for this chapter (this assumes you’ve already installed them). In this chapter we introduce some new packages:
- The
tidyverse“umbrella” package. Recall from our discussion in Section 5.4.1 that loading thetidyversepackage by runninglibrary(tidyverse)loads the following commonly used data science packages all at once:ggplot2for data visualizationdplyrfor data wranglingtidyrfor converting data to “tidy” formatreadrfor importing spreadsheet data into R- As well as the more advanced
purrr,tibble,stringr, andforcatspackages
- The
moderndivepackage of datasets and functions for tidyverse-friendly introductory linear regression. - The
skimrpackage which provides a simple to use summary function that can be used with pipes and displays nicely in the console.
If needed, read Section 2.3 for information on how to install and load R packages.
library(tidyverse)
library(moderndive)
library(skimr)
library(gapminder)6.1 One numerical explanatory variable
Why do some professors and instructors at universities and colleges receive high teaching evaluations from students while others don’t? Are there differences in teaching evaluations between instructors of different demographic groups? Could there be an impact due to student biases? These are all questions that are of interest to university/college administrators, as teaching evaluations are among the many criteria considered in determining which instructors and professors get promoted.
Researchers at the University of Texas in Austin, Texas (UT Austin) tried to answer the following research question: what factors can explain differences in instructor’s teaching evaluation scores? To this end, they collected instructor and course information on 463 courses. A full description of the study can be found at openintro.org.
In this section, we’ll keep things simple for now and try to explain differences in instructor teaching scores as a function of one numerical variable: the instructor’s “beauty score”; we’ll describe how this score was computed shortly. Could it be that instructors with higher beauty scores also have higher teaching evaluations? Could it be instead that instructors with higher beauty scores tend to have lower teaching evaluations? Or could it be there is no relationship between beauty score and teaching evaluations? We’ll answer these questions by modeling the relationship between teaching scores and “beauty scores” using simple linear regression where we have:
- A numerical outcome variable \(y\), the instructor’s teaching score and
- A single numerical explanatory variable \(x\), the instructor’s beauty score.
6.1.1 Exploratory data analysis
The data on the 463 courses at the UT Austin can be found in the evals data frame included in the moderndive package. However, to keep things simple, let’s select() only the subset of the variables we’ll consider in this chapter, and save this data in a new data frame called eval_ch6:
evals_ch6 <- evals %>%
select(ID, score, bty_avg, age)A crucial step before doing any kind of analysis or modeling is performing an exploratory data analysis, or EDA for short. Exploratory data analysis gives you a sense of the distributions of the individual variables in your data, whether there are outliers and/or missing values, and most importantly help inform how to build your model. Here are three common steps in an exploratory data analysis.
- Most crucially: Looking at the raw data values.
- Computing summary statistics, like means, medians, and interquartile ranges.
- Creating data visualizations.
Let’s perform the first common step in an exploratory data analysis: looking at the raw data values. Unfortunately, many analysts ignore the first step. Because this step seems so trivial, many analysts often ignore it. However, getting an early sense of what your raw data looks like can often prevent many larger issues down the road. You can do this by using RStudio’s spreadsheet viewer or by using the glimpse() function as introduced in Section 2.4.3 on exploring data frames:
glimpse(evals_ch6)Observations: 463
Variables: 4
$ ID <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18…
$ score <dbl> 4.7, 4.1, 3.9, 4.8, 4.6, 4.3, 2.8, 4.1, 3.4, 4.5, 3.8, 4.5, 4…
$ bty_avg <dbl> 5.00, 5.00, 5.00, 5.00, 3.00, 3.00, 3.00, 3.33, 3.33, 3.17, 3…
$ age <int> 36, 36, 36, 36, 59, 59, 59, 51, 51, 40, 40, 40, 40, 40, 40, 4…Observe that Observations: 463 indicates that there are 463 rows/observations in evals_ch6, where each row corresponds to one observed course at UT Austin. It is important to note that the observational unit are individual courses and not individual instructors. The observational unit is the thing that is being measured or with which the variables provide characteristics for. Since instructors often teach more than one course in an academic year, the same instructor can often appear more than once in the data. Hence there are fewer than 463 unique instructors being represented in evals_ch6. We’ll revisit this idea in Chapter 11, when we talk about the “independence assumption” for inference for regression.
While a full description of all the variables included in evals can be found at openintro.org and by reading the associated help file by running ?evals in the Console, let’s fully describe the 4 variables we selected in evals_ch6:
ID: An identification variable used to distinguish between the 1 through 463 courses in the dataset.score: A numerical variable of the course instructor’s average teaching score, where the average is computed from the evaluation scores from all students in that course. Teaching scores of 1 are lowest and 5 are highest. This is the outcome variable \(y\) of interest.bty_avg: A numerical variable of the course instructor’s average “beauty” score, where the average is computed from a separate panel of 6 students. “Beauty” scores of 1 are lowest and 10 are highest. This is the explanatory variable \(x\) of interest.age: A numerical variable of the course instructor’s age. This will be another explanatory variable \(x\) we’ll study later.
An alternative way to look at the raw data values is by choosing a random sample of the courses, i.e. rows in evals_ch6 by piping it into the sample_n() function from the dplyr package. Here we set the size argument to be 5, indicating that we want a random sample of 5 rows. We display the results Table 6.1. Note due to the random nature of the sampling, you will likely end up with a different subset of 5 rows.
evals_ch6 %>%
sample_n(size = 5)| ID | score | bty_avg | age |
|---|---|---|---|
| 129 | 3.7 | 3.00 | 62 |
| 109 | 4.7 | 4.33 | 46 |
| 28 | 4.8 | 5.50 | 62 |
| 434 | 2.8 | 2.00 | 62 |
| 330 | 4.0 | 2.33 | 64 |
Now that we’ve looked at the raw values in our evals_ch6 data frame and obtained a sense of the data, let’s move on to next common step in an exploratory data analysis: computing summary statistics. Let’s start by computing the mean and median of our numerical outcome variable score and our numerical explanatory variable bty_avg beauty score:
evals_ch6 %>%
summarize(mean_bty_avg = mean(bty_avg), mean_score = mean(score),
median_bty_avg = median(bty_avg), median_score = median(score))# A tibble: 1 x 4
mean_bty_avg mean_score median_bty_avg median_score
<dbl> <dbl> <dbl> <dbl>
1 4.42 4.17 4.33 4.3However, what if we want other summary statistics as well, such as the standard deviation (a measure of spread), the minimum and maximum values, and various percentiles? Typing out all these summary statistic functions in the earlier summarize() would be long and tedious. Instead, let’s using the very convenient skim() function from the skimr package. This function takes in a data frame, “skims” it, and returns commonly used summary statistics. Let’s take our evals_ch6 data frame, select() only the outcome and explanatory variables teaching score and bty_avg, and pipe it into the skim() function:
evals_ch6 %>%
select(score, bty_avg) %>%
skim()Skim summary statistics
n obs: 463
n variables: 2
── Variable type:numeric ───────────────────────────────────────────────────────
variable missing complete n mean sd p0 p25 p50 p75 p100
bty_avg 0 463 463 4.42 1.53 1.67 3.17 4.33 5.5 8.17
score 0 463 463 4.17 0.54 2.3 3.8 4.3 4.6 5 (Note that for formatting purposes, the inline histogram that is usually printed with skim() has been removed.)
For our two numerical variables teaching score and beauty score bty_avg it returns:
missing: the number of missing valuescomplete: the number of non-missing or complete valuesn: the total number of valuesmean: the mean AKA averagesd: the standard deviationp0: the 0th percentile: the value at which 0% of observations are smaller than it AKA the minimum valuep25: the 25th percentile: the value at which 25% of observations are smaller than it AKA the 1st quartilep50: the 50th percentile: the value at which 50% of observations are smaller than it AKA the 2nd quartile and more commonly the medianp75: the 75th percentile: the value at which 75% of observations are smaller than it AKA the 3rd quartilep100: the 100th percentile: the value at which 100% of observations are smaller than it AKA the maximum value
Looking at the earlier output we get an idea of how the values of both variables distribute. For example, the mean teaching score was 4.17 out of 5 whereas the mean beauty score was 4.42 out of 10. Furthermore, the middle 50% of teaching scores were between 3.80 and 4.6 (the first and third quartiles) whereas the middle 50% of beauty scores were between 3.17 and 5.5 out of 10.
However, the skim() function only returns what are known as univariate summary statistics: functions that take a single variable and return some summary of that variable. However, there also exist bivariate summary statistics: functions that take in two variables and return some summary of those two variables. In particular, when the two variables are numerical we can compute the correlation coefficient. Generally speaking, coefficients are quantitative expressions of a specific property of a phenomenon. A correlation coefficient is a quantitative expression of the strength of the linear relationship between two numerical variables whose value range between -1 and 1 where:
- -1 indicates a perfect negative relationship: As the value of one variable goes up, the value of the other variable tends to go down.
- 0 indicates no relationship: The values of both variables go up/down independently of each other.
- +1 indicates a perfect positive relationship: As the value of one variable goes up, the value of the other variable tends to go up as well.
Figure 6.1 gives examples of 9 different correlation coefficient values for hypothetical numerical variables \(x\) and \(y\). For example, observe that for a correlation coefficient of -0.75 there is still a negative linear relationship between \(x\) and \(y\), it is not as strong as the negative linear relationship between \(x\) and \(y\) when the correlation coefficient is -0.9 or -1.
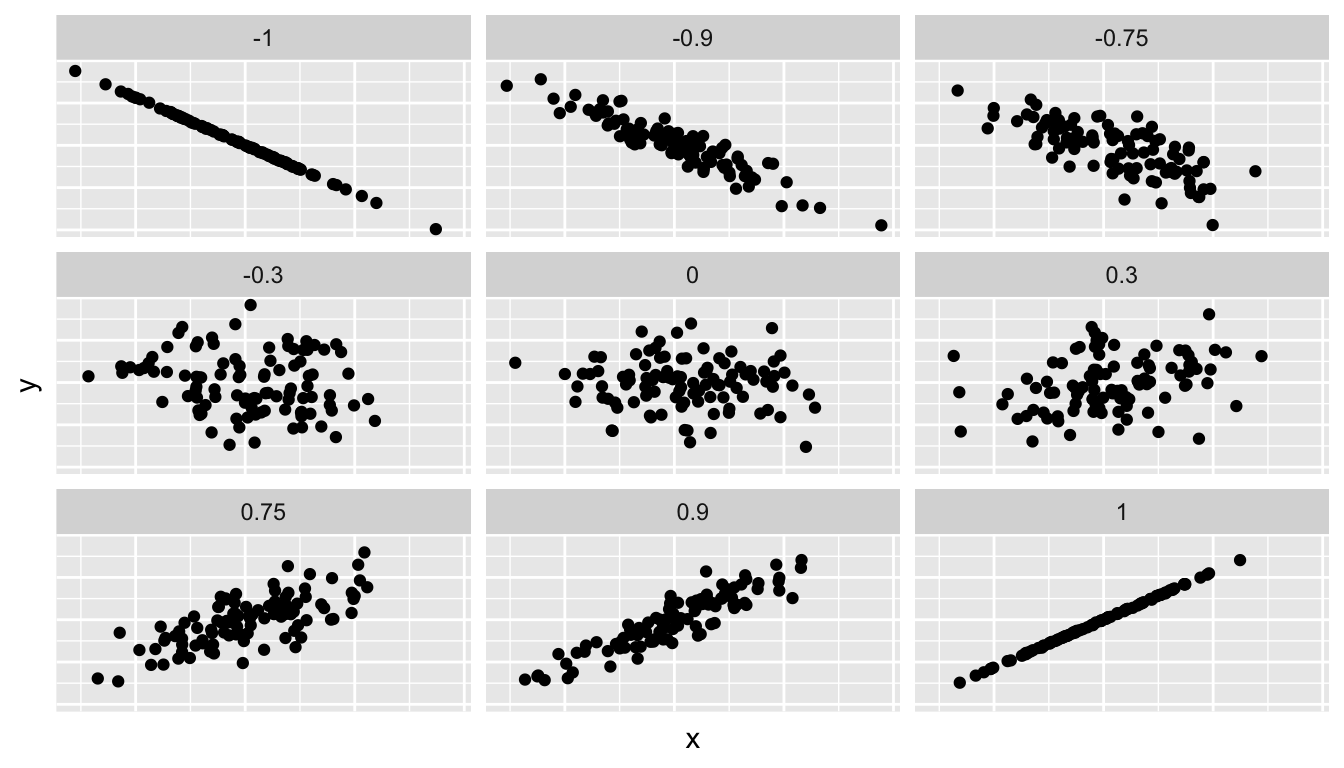
FIGURE 6.1: Different correlation coefficients.
The correlation coefficient can be computed using the get_correlation() function in the moderndive package, where in this case the inputs to the function are the two numerical variables from which we want to calculate the correlation coefficient. We place the name of the response variable on the left hand side of the ~ and the explanatory variable on the right hand side of the “tilde” in R’s formula notation. We will use this same “formula” syntax with regression later in this chapter.
evals_ch6 %>%
get_correlation(formula = score ~ bty_avg)# A tibble: 1 x 1
correlation
<dbl>
1 0.187An alternative way to compute the correlation coefficient is to use the cor() function within a summarize():
evals_ch6 %>%
summarize(correlation = cor(score, bty_avg))# A tibble: 1 x 1
correlation
<dbl>
1 0.187In our case, the correlation coefficient of 0.187 indicates that the relationship between teaching evaluation score and beauty average is “weakly positive.” There is a certain amount of subjectivity in interpreting correlation coefficients, especially those that aren’t close to the extreme values of -1, 0, and 1. To develop intuition in interpreting correlation coefficients see play the “Guess the Correlation” 1980’s style video game in Subsection 6.4.1.
Let’s now perform the last of the three common steps in an exploratory data analysis: creating data visualizations. Since both the score and bty_avg variables are numerical, a scatterplot is an appropriate graph to visualize this data. Let’s do this using geom_point() and display the result in Figure 6.2. Furthermore, let’s highlight the 6 points in the top right of the visualization in orange.
ggplot(evals_ch6, aes(x = bty_avg, y = score)) +
geom_point() +
labs(x = "Beauty Score", y = "Teaching Score",
title = "Scatterplot of relationship of teaching and beauty scores")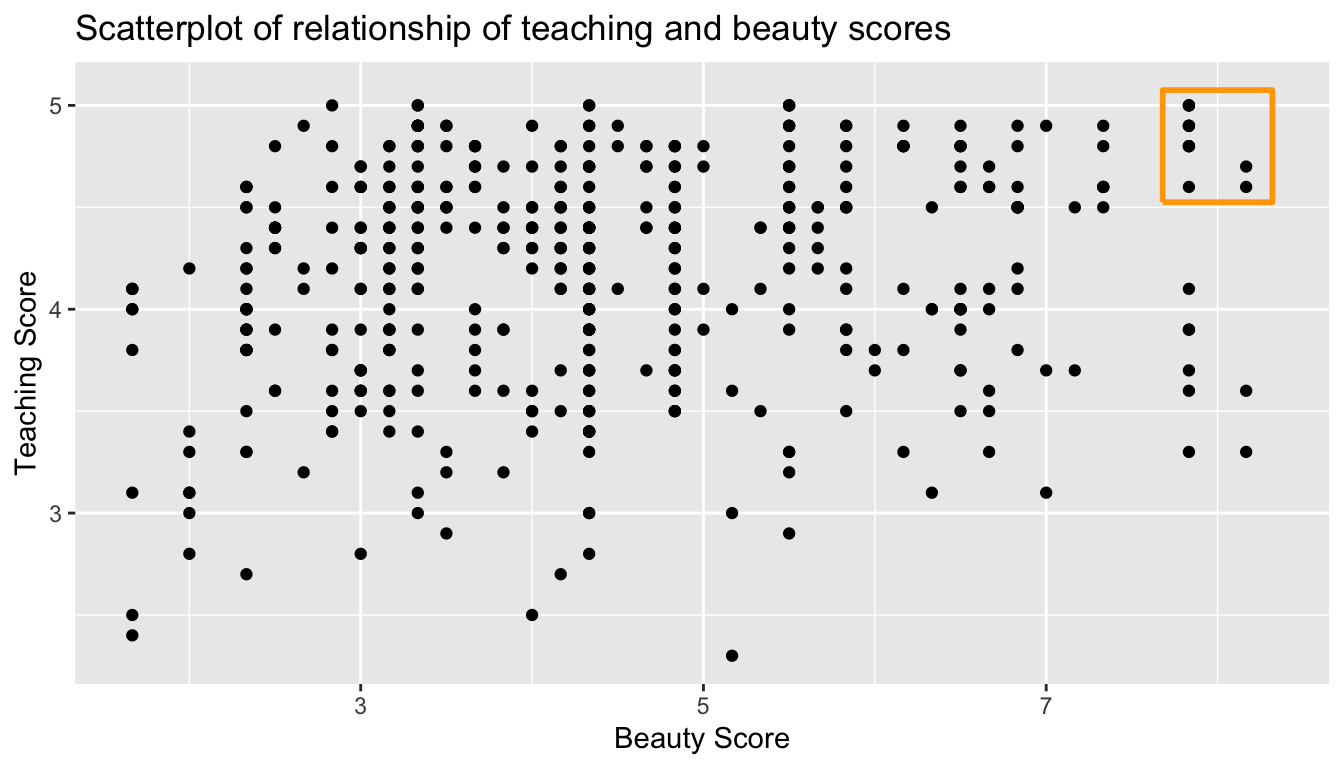
FIGURE 6.2: Instructor evaluation scores at UT Austin.
Observe the following:
- Most “beauty” scores lie between 2 and 8.
- Most teaching scores lie between 3 and 5.
- While opinions may vary, it is our opinion that the relationship between teaching score and beauty score appears weakly positive. This is consistent with our earlier computed correlation coefficient of 0.187.
Furthermore, there appear to be 6 points in the top-right of this plot highlighted in the orange box. However, this is not the case as this plot suffers from overplotting. Recall from Subsection 3.3.2 that overplotting occurs when several points are stacked directly on top of each other, thereby obscuring their number. So while it may appear that there are only 6 points in the orange box, there are actually more. This fact is only apparent when using geom_jitter() in place of geom_point(). We display the resulting plot in Figure 6.3 along with the same orange box as in Figure 6.3.
ggplot(evals_ch6, aes(x = bty_avg, y = score)) +
geom_jitter() +
labs(x = "Beauty Score", y = "Teaching Score",
title = "Scatterplot of relationship of teaching and beauty scores")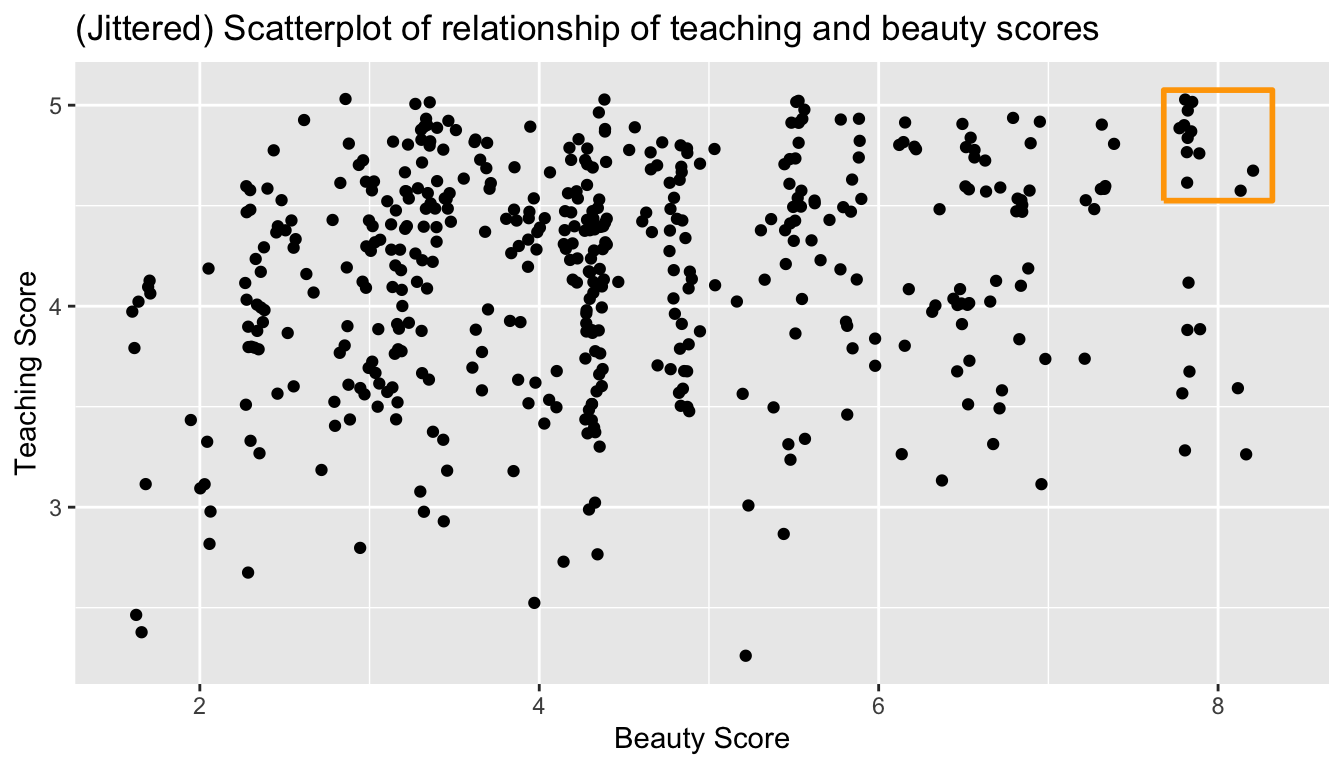
FIGURE 6.3: Instructor evaluation scores at UT Austin.
It is now apparent that there are 12 points in the area highlighted in orange and not 6 as originally suggested in Figure 6.2. Recall from Section 3.3.2 on overplotting that jittering adds a little random “nudge” to each of the points to break up these ties. Furthermore, jittering is strictly a visualization tool; it does not alter the original values in the data frame evals_ch6. To keep things simple going forward however, we’ll only present regular scatterplots rather than their jittered counterparts.
Let’s build on the unjittered scatterplot in Figure 6.2 by adding a “best-fitting” line; of all possible lines we can draw on this scatterplot, its the line that “best” fits through the cloud of points. We do this by adding a new geom_smooth(method = "lm", se = FALSE) layer to the ggplot() code that created the scatterplot in Figure 6.2. The method = lm argument sets the line to be a “linear model” while the se = FALSE argument suppresses “standard error” uncertainty bars.
ggplot(evals_ch6, aes(x = bty_avg, y = score)) +
geom_point() +
labs(x = "Beauty Score", y = "Teaching Score",
title = "Relationship between teaching and beauty scores") +
geom_smooth(method = "lm", se = FALSE)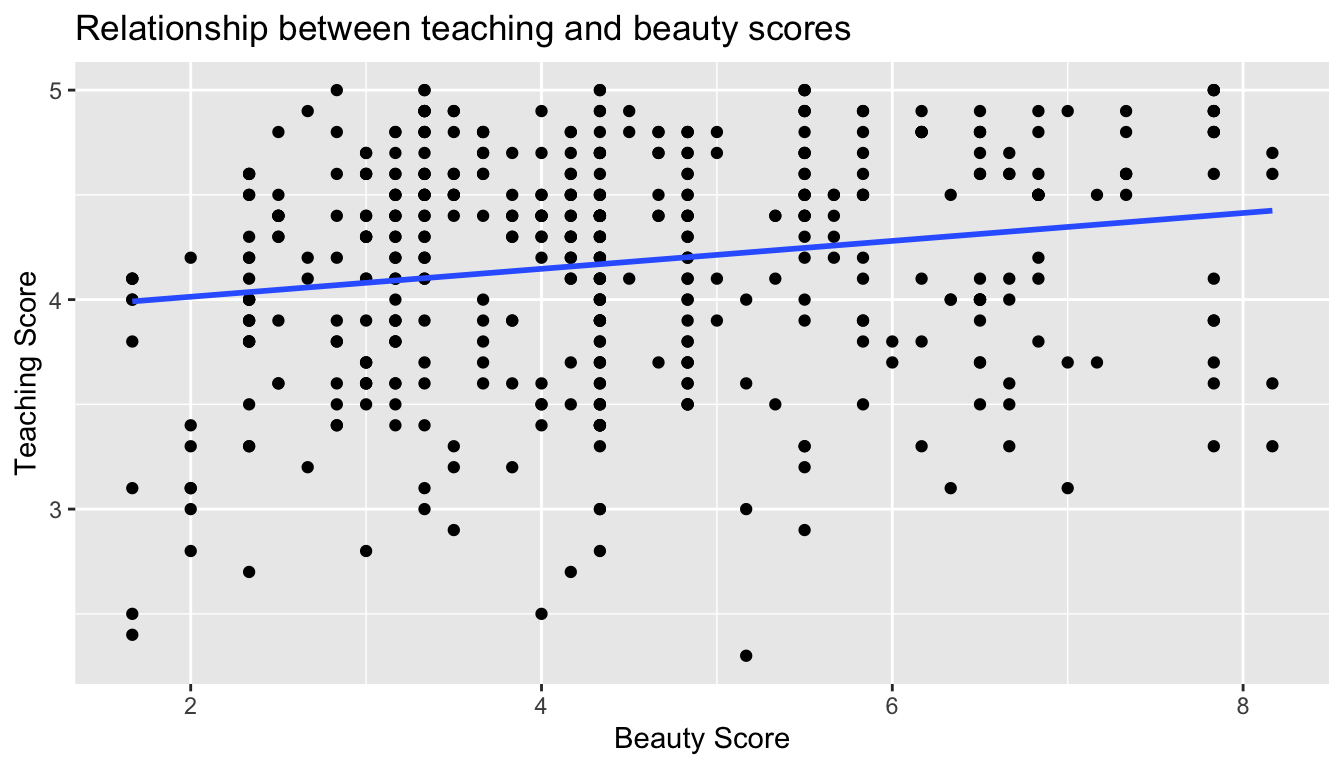
FIGURE 6.4: Regression line.
The blue line in the resulting Figure 6.4 is called a “regression line”. The regression line is a visual summary of the relationship between two numerical variables, in our case the outcome variable score and the explanatory variable bty_avg. The positive slope of the blue line is consistent with our earlier observed correlation coefficient of 0.187 suggesting that there is a positive relationship between these two variables: as instructors have higher beauty scores so also do they have receive higher teaching evaluations. We’ll see later however that while the correlation coefficient and the slope of a regression line always have the same sign (positive or negative), they do not necessarily have the same value.
Furthermore, a regression line is “best” fitting in that it minimizes some mathematical criteria. We present this mathematical criteria in Subsection 6.3.2, but we suggest you read this subsection only after reading the rest of this section on regression with one numerical explanatory variable.
Learning check
(LC6.1) Conduct a new exploratory data analysis with the same outcome variable \(y\) being score but with age as the new explanatory variable \(x\). Remember, this involves three things:
- Looking at the raw data values.
- Computing summary statistics.
- Creating data visualizations.
What can you say about the relationship between age and teaching scores based on this exploration?
6.1.2 Simple linear regression
You may recall from secondary school / high school algebra, the equation of a line is \(y = a + b\cdot x\) which is defined by two coefficients \(a\) and \(b\) (recall from earlier that coefficients are “quantitative expressions of a specific property of a phenomenon”): the intercept coefficient \(a\) i.e. the value of \(y\) when \(x = 0\) and the slope coefficient \(b\) for x i.e. the increase in \(y\) for every increase of one in \(x\).
However, when defining a regression line like the blue regression line in Figure 6.4, we use slightly different notation: the equation of the regression line is \(\widehat{y} = b_0 + b_1 \cdot x\) where the intercept coefficient is \(b_0\) i.e. the value of \(\widehat{y}\) when \(x=0\). The slope coefficient is \(b_1\) for x i.e. the increase in \(\widehat{y}\) for every increase of one in \(x\). Why do we put a “hat” on top of the \(y\)? It’s a form of notation commonly used in regression to indicate that we have a “fitted value”, or the value of \(y\) on the regression line for a given \(x\) value; we’ll discussion this more in the upcoming Subsection 6.1.3.
We know that the blue regression line in Figure 6.4 has a positive slope \(b_1\) corresponding to our explanatory \(x\) variable bty_avg. Why? Because as instructors have higher bty_avg scores, so also do they tend to have higher teaching evaluation scores. However, what is the specific numerical value of the slope \(b_1\)? What about the intercept \(b_0\)? Let’s compute these two values by hand, but rather let’s use a computer!
We can obtain the intercept \(b_0\) and slope \(b_1\) for btg_avg by outputting a linear regression table. This is done in two steps:
- We first “fit” the linear regression model using the
lm()function and save it inscore_model. - We get the regression table by applying the
get_regression_table()from themoderndivepackage toscore_model.
# Fit regression model:
score_model <- lm(score ~ bty_avg, data = evals_ch6)
# Get regression table:
get_regression_table(score_model)| term | estimate | std_error | statistic | p_value | lower_ci | upper_ci |
|---|---|---|---|---|---|---|
| intercept | 3.880 | 0.076 | 50.96 | 0 | 3.731 | 4.030 |
| bty_avg | 0.067 | 0.016 | 4.09 | 0 | 0.035 | 0.099 |
Let’s first focus on interpreting the regression table output in Table 6.2 and then later we’ll revisit the code that produced it. In the estimate column of Table 6.2 are the intercept \(b_0\) = 3.88 and the slope \(b_1\) = 0.067 for bty_avg and thus the equation of the blue regression line in Figure 6.4 follows (note that the \(\cdot\) symbol is equivalent to the \(\times\) “multiply by” mathematical symbol; we use the \(\cdot\) symbol in this book as it is a little cleaner):
\[ \begin{aligned} \widehat{y} &= b_0 + b_1 \cdot x\\ \widehat{\text{score}} &= b_0 + b_{\text{bty}\_\text{avg}} \cdot\text{bty}\_\text{avg}\\ &= 3.880 + 0.067\cdot\text{bty}\_\text{avg} \end{aligned} \]
The intercept \(b_0\) = 3.8803 is the value average teaching score \(\widehat{y}\) = \(\widehat{\text{score}}\) for those courses where the instructor had a beauty score bty_avg of 0. In other words, it’s where the line intersects the \(y\) axis when \(x\) = 0. Note however that while the intercept of the regression line has a mathematical interpretation, it has no practical interpretation since observing a bty_avg of 0 is impossible; it is the average of six panelists’ beauty score ranging from 1 to 10. Furthermore, looking at the scatterplot with the regression line in Figure 6.4, no instructors had a beauty score anywhere near 0.
Of greater interest is the slope \(b_1\) = \(b_{\text{bty avg}}\) for bty_avg of +0.067, as this summarizes the relationship between teaching score and beauty score. Note that the sign is positive suggesting a positive relationship between these two variables, meaning teachers with higher beauty scores also tend to have higher teaching scores. Recall from earlier that the correlation coefficient is 0.187: they both have the same positive sign, but have a different value. Recall further that the correlation’s interpretation is the “strength of linear association”. The slope’s interpretation is a little different:
For every increase of 1 unit in
bty_avg, there is an associated increase of, on average, 0.0666 units ofscore.
We only state that there is an associated increase and not necessarily a causal increase. For example, perhaps it’s not that higher beauty scores directly cause higher teaching scores per se. Instead it could be that individuals from wealthier backgrounds tend to have stronger educational backgrounds and hence have higher teaching scores, but that these wealthy individuals also have higher beauty scores. In other words, just because two variables are strongly associated doesn’t mean that one necessarily causes the other. This is summed up in the often quoted phrase “correlation is not necessarily causation.” We discuss this idea further in Subsection 6.3.1.
Furthermore, we say that this associated increase is on average 0.067 units of teaching score because you might have two instructors whose bty_avg score differ by 1 unit, but their difference in teaching scores isn’t necessarily 0.067. What the slope of 0.067 is across all courses, the average difference in teaching score between two instructors whose beauty scores differ by one is 0.067.
Now that we’ve learned how to compute the equation for the blue regression line in Figure 6.4 using the values in the estimate column of Table 6.2 and how to interpret the resulting the intercept and slope, let’s revisit the code that generated this table:
# Fit regression model:
score_model <- lm(score ~ bty_avg, data = evals_ch6)
# Get regression table:
get_regression_table(score_model)First, we “fit” the linear regression model to the data using the lm() function and save this to score_model. When we say “fit”, we mean “find the best fitting line to this data.” lm() stands for “linear model” and is used as follows: lm(y ~ x, data = data_frame_name) where:
yis the outcome variable, followed by a tilde (~); this is likely the key to the left of the “1” key on your keyboard. In our case,yis set toscore.xis the explanatory variable. In our case,xis set tobty_avg.- The combination of
y ~ xis called a model formula (note the order ofyandx). In our case, the model formula isscore ~ bty_avg. We saw such model formulas earlier as well when we computed the correlation coefficient using theget_correlation()function in Subsection 6.1.1. data_frame_nameis the name of the data frame that contains the variablesyandx. In our case,data_frame_nameis theevals_ch6data frame.
Second, we take the saved model in score_model and apply the get_regression_table() function from the moderndive package to it to obtain the regression table in Table 6.2. This function is an example of what’s known as a wrapper function in computer programming, which takes other pre-existing functions and “wraps” them into a single function that hides its inner workings. This concept is illustrated in Figure 6.5.
FIGURE 6.5: The concept of a wrapper function.
So all you need to worry about is the what the inputs look like and what the outputs look like; you leave all the other details “under the hood of the car.” In our regression modeling example, the get_regression_table() function takes as input a saved lm() linear regression and returns as output a data frame of the regression table with information on the intercept and slope of the regression line. If you’re interested in learning more about the get_regression_table() function’s design and inner-workings, check out Subsection 6.3.3.
Lastly, you might be wondering what remaining 5 columns in Table 6.2 are: std_error, statistic, p_value, lower_ci and upper_ci? They are the “standard error”, “test statistic”, “p-value”, “lower 95% confidence interval bound”, and “upper 95% confidence interval bound.” They tell us about both the statistical significance and practical significance of our results. You can think of this loosely as the “meaningfulness” of our results from a statistical perspective. We are going to put aside these ideas for now and revisit them in Chapter 11 on (statistical) inference for regression, after we’ve had a chance to cover:
- Standard errors in Chapter 8
- Confidence intervals in Chapter 9
- Hypothesis testing and p-values in Chapter 10
Learning check
(LC6.2) Fit a new simple linear regression using lm(score ~ age, data = evals_ch6) where age is the new explanatory variable \(x\). Get information about the “best-fitting” line from the regression table by applying the get_regression_table() function. How do the regression results match up with the results from your earlier exploratory data analysis?
6.1.3 Observed/fitted values and residuals
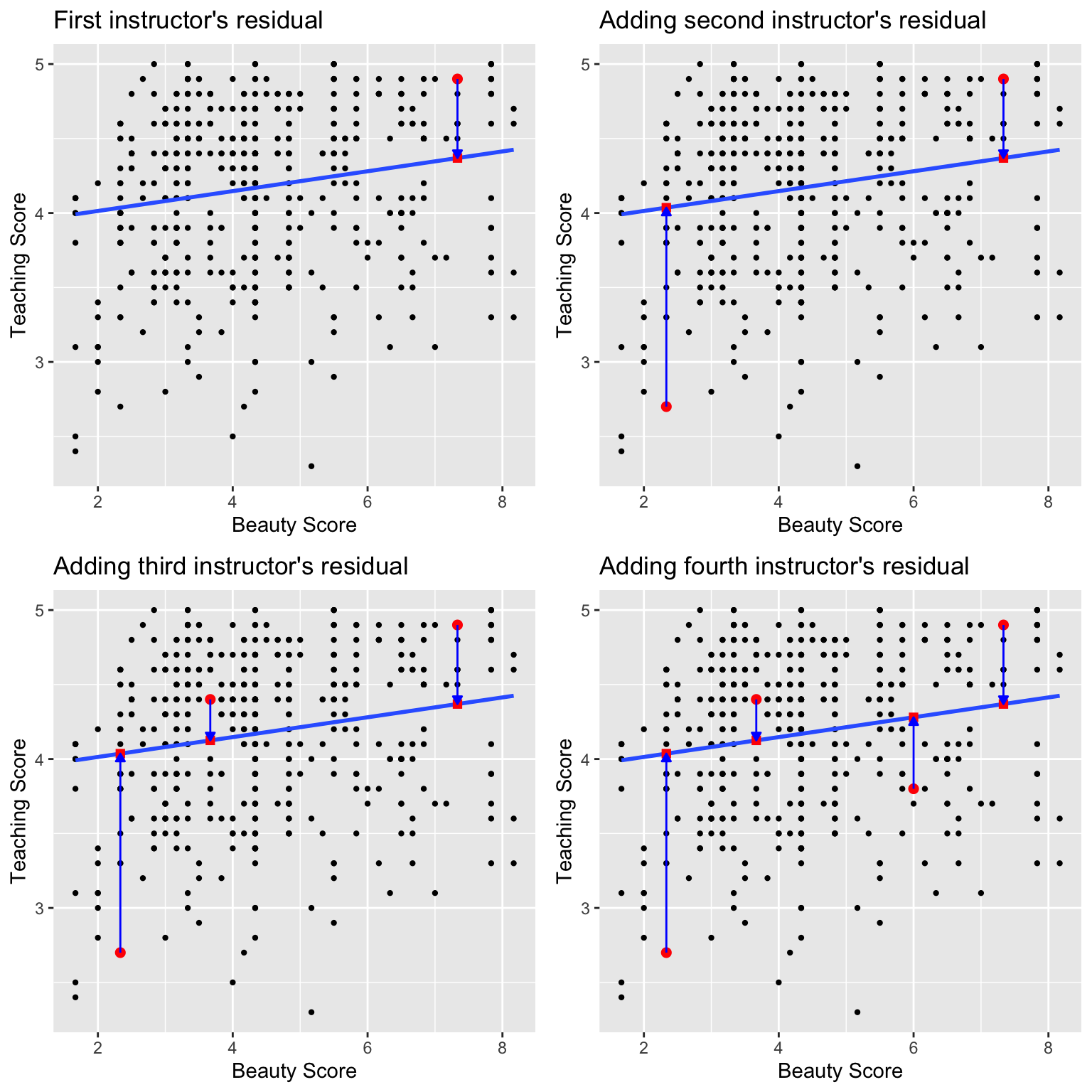
We just saw how to get the value of the intercept and the slope of a regression line from the estimate column of a regression table generated by get_regression_table(). Now instead say we want information on individual observations. For example, let’s focus on 21st of the 463 courses in the evals_ch6 data frame in Table 6.3:
| ID | score | bty_avg | age |
|---|---|---|---|
| 21 | 4.9 | 7.33 | 31 |
What is the value \(\widehat{y}\) on the blue line regression line corresponding to this instructor’s bty_avg beauty score of 7.333? In Figure 6.6 we mark three values corresponding to the instructor for this 21st course:
- Red circle: The observed value \(y\) = 4.9 is this course’s instructor’s actual teaching score.
- Red square: The fitted value \(\widehat{y}\) is value on the regression line for \(x\) =
bty_avg= 7.333. This value is computed using the intercept and slope in the previous regression table:
\[\widehat{y} = b_0 + b_1 \cdot x = 3.88 + 0.067 \cdot 7.333 = 4.369\]
- Blue arrow: The length of this arrow is the residual and is computed by subtracting the fitted value \(\widehat{y}\) from the observed value \(y\). The residual can be thought of as the error or “lack of fit”. In the case of this course’s instructor, it is \(y - \widehat{y}\) = 4.9 - 4.369 = 0.531.
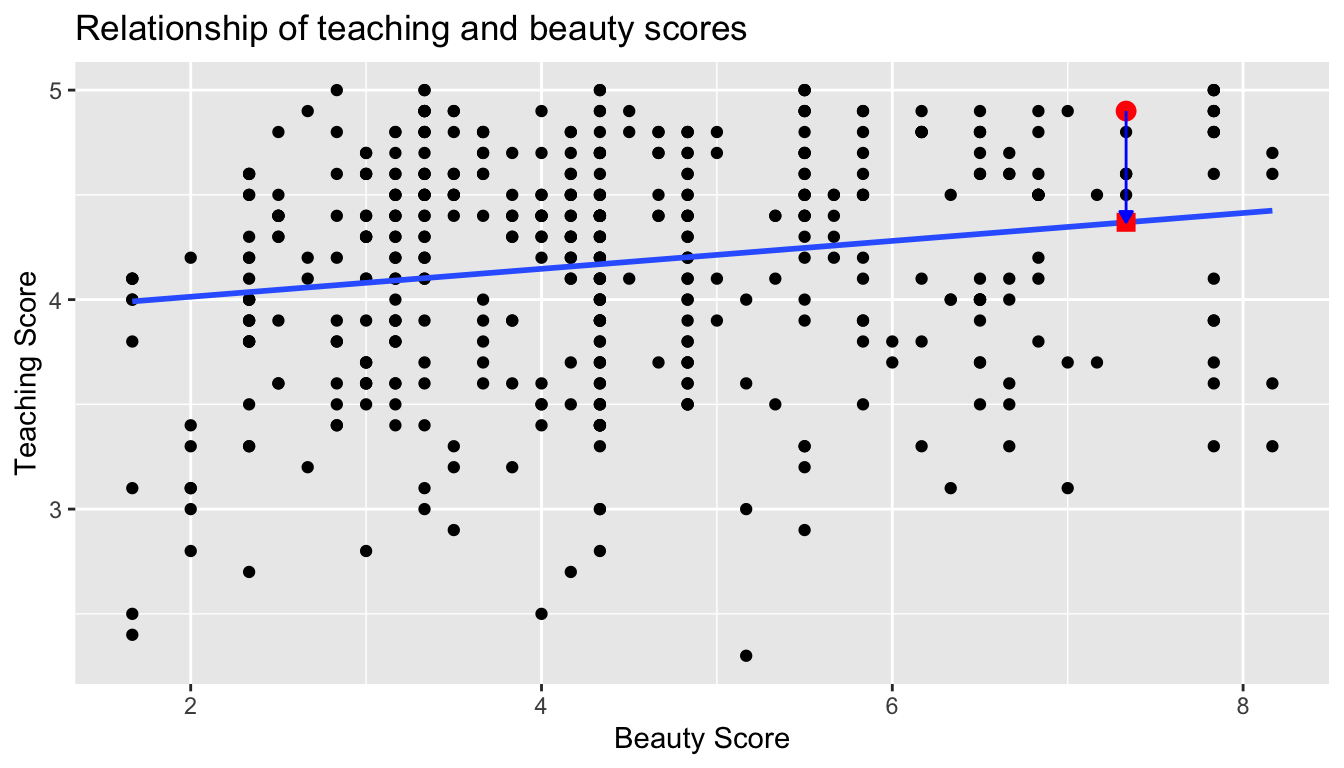
FIGURE 6.6: Example of observed value, fitted value, and residual.
Now say we want to compute both the
- the fitted value \(\widehat{y} = b_0 + b_1 \cdot x\) and
- the residual \(y - \widehat{y}\)
not only for the instructor of the 21st course, but for all 463 courses in the study? Recall that each course corresponds to one of the 463 rows in the evals_ch6 data frame and also one of the 463 points in the regression plot in Figure 6.6.
We could repeat the previous calculations we performed by hand 463 times, but that would be tedious and time consuming. Instead, let’s use the computer using the get_regression_points() function included in the moderndive package. Just like the get_regression_table() function, the get_regression_points() function is a “wrapper” function; however it returns a different output. Let’s apply the get_regression_points() function to score_model, which is where we saved our lm() model in the previous section. In Table 6.4 we present only the results of the 21st through 24th courses for brevity’s sake.
regression_points <- get_regression_points(score_model)
regression_points| ID | score | bty_avg | score_hat | residual |
|---|---|---|---|---|
| 21 | 4.9 | 7.33 | 4.37 | 0.531 |
| 22 | 4.6 | 7.33 | 4.37 | 0.231 |
| 23 | 4.5 | 7.33 | 4.37 | 0.131 |
| 24 | 4.4 | 5.50 | 4.25 | 0.153 |
Let’s inspect the individual columns and match them with the elements of Figure 6.6:
- The
scorecolumn represents the observed outcome variable \(y\) i.e. the y-position of the 463 black points. - The
bty_avgcolumn represents the values of the explanatory variable \(x\) i.e. the x-position of the 463 black points. - The
score_hatcolumn represents the fitted values \(\widehat{y}\) i.e. the corresponding value on the blue regression line for the 463 \(x\) values. - The
residualcolumn represents the residuals \(y - \widehat{y}\) i.e the 463 vertical distances between the 463 black points and the blue regression line.
Just as we did for the instructor of the 21st course in the evals_ch6 dataset (in the first row of the table above), let’s repeat the calculations for the instructor of the 24th course (in the fourth row of Table 6.4 above):
score= 4.4 is the observed teachingscore\(y\) for this course’s instructor.bty_avg= 5.50 is the value of the explanatory variablebty_avg\(x\) for this course’s instructor.score_hat= 4.25 = 3.88 + 0.067 \(\cdot\) 5.50 is the fitted value \(\widehat{y}\) on the blue regression line for this course’s instructor.residual= 0.153 = 4.4 - 4.25 is the value of the residual for this instructor. In other words, the model was off by 0.153 teaching score units for this course’s instructor.
At this point we suggest you read Section 6.3.2 in which we define what we mean by “best” for “best-fitting” regression lines: it is the line that minimizes the sum of squared residuals.
Learning check
(LC6.3) Generate a data frame of the residuals of the model where you used age as the explanatory \(x\) variable.
6.2 One categorical explanatory variable
It’s an unfortunate truth that life expectancy is not the same across all countries in the world. International development agencies are very interested in studying these differences in life expectancy in the hopes of identifying where governments should allocate resources to address this problem. In this section, we’ll explore differences in life expectancy in two ways:
- Differences between continents: Are there significant differences in average life expectancy between the five continents of the world: Africa, the Americas, Asia, Europe, and Oceania?
- Differences within continents: How does life expectancy vary within the world’s five continents? For example, is the spread of life expectancy among the countries of Africa larger than the spread of life expectancy among the countries of Asia?
To answer such questions, we’ll use the gapminder data frame included in the gapminder package. This dataset has international development statistics such as life expectancy, GDP per capita, and population for 142 countries for 5-year intervals between 1952 and 2007. Recall we visualized this data in Figure 3.1 in Subsection 3.1.2 on the “Grammar of Graphics”.
We’ll use this data for basic linear regression again but now using an explanatory variable \(x\) that is categorical, as opposed to the numerical explanatory variable model we saw in Section 6.1 on instructor teaching and beauty scores. In this section, we’ll model the relationship between
- A numerical outcome variable \(y\), a country’s life expectancy and
- A single categorical explanatory variable \(x\), the continent the country is a part of.
When the explanatory variable \(x\) is categorical, the concept of a “best-fitting” regression line is a little different than the one we saw previously in Section 6.1 where the explanatory variable \(x\) was numerical. We’ll study these differences shortly in Subsection 6.2.2, but first we conduct our exploratory data analysis.
6.2.1 Exploratory data analysis
The data on the 142 countries can be found in the gapminder data frame included in the gapminder package. However, to keep things simple, let’s filter() for only observations/rows corresponding to the year 2007, select() only the subset of the variables we’ll consider in this chapter, and save this data in a new data frame called gapminder2007:
library(gapminder)
gapminder2007 <- gapminder %>%
filter(year == 2007) %>%
select(country, lifeExp, continent, gdpPercap)Recall from Section 6.1.1 that there are three common steps in an exploratory data analysis:
- Most crucially: Looking at the raw data values.
- Computing summary statistics, like means, medians, and interquartile ranges.
- Creating data visualizations.
Let’s perform the first common step in an exploratory data analysis: looking at the raw data values. You can do this by using RStudio’s spreadsheet viewer or by using the glimpse() command as introduced in Section 2.4.3 on exploring data frames:
glimpse(gapminder2007)Observations: 142
Variables: 4
$ country <fct> Afghanistan, Albania, Algeria, Angola, Argentina, Australia…
$ lifeExp <dbl> 43.8, 76.4, 72.3, 42.7, 75.3, 81.2, 79.8, 75.6, 64.1, 79.4,…
$ continent <fct> Asia, Europe, Africa, Africa, Americas, Oceania, Europe, As…
$ gdpPercap <dbl> 975, 5937, 6223, 4797, 12779, 34435, 36126, 29796, 1391, 33…Observe that Observations: 142 indicates that there are 142 rows/observations in gapminder2007, where each row corresponds to one country. In other words, the observational unit are individual countries. Furthermore, observe that the variable continent is of type <fct>, which stands for “factor,” which is R’s way of encoding categorical variables.
While a full description of all the variables included in gapminder can be found by reading the associated help file by running ?gapminder in the Console, let’s fully describe the 4 variables we selected in gapminder2007:
country: An identification variable used to distinguish the 142 countries in the dataset.lifeExp: A numerical variable of that country’s life expectancy at birth. This is the outcome variable \(y\) of interest.continent: A categorical variable with 5 levels i.e. possible categories: Africa, Asia, Americas, Europe, and Oceania. This is the explanatory variable \(x\) of interest.gdpPercap: A numerical variable of that country’s GDP per capita in US inflation-adjusted dollars that we’ll use as another outcome variable \(y\) in the Learning Check at the end of this section.
Furthermore, let’s look at a random sample of 5 out of the 142 countries in Table 6.5. Note due to the random nature of the sampling, you will likely end up with a different subset of 5 rows.
gapminder2007 %>%
sample_n(size = 5)| country | lifeExp | continent | gdpPercap |
|---|---|---|---|
| Togo | 58.4 | Africa | 883 |
| Sao Tome and Principe | 65.5 | Africa | 1598 |
| Congo, Dem. Rep. | 46.5 | Africa | 278 |
| Lesotho | 42.6 | Africa | 1569 |
| Bulgaria | 73.0 | Europe | 10681 |
Now that we’ve looked at the raw values in our gapminder2007 data frame and obtained a sense of the data, let’s move on to next common step in an exploratory data analysis: computing summary statistics. Let’s once again apply the skim() function from the skimr package. Recall from our previous EDA that this function takes in a data frame, “skims” it, and returns commonly used summary statistics. Let’s take our gapminder2007 data frame, select() only the outcome and explanatory variables lifeExp and continent, and pipe it into the skim() function:
gapminder2007 %>%
select(lifeExp, continent) %>%
skim()Skim summary statistics
n obs: 142
n variables: 2
── Variable type:factor ────────────────────────────────────────────────────────
variable missing complete n n_unique top_counts ordered
continent 0 142 142 5 Afr: 52, Asi: 33, Eur: 30, Ame: 25 FALSE
── Variable type:numeric ───────────────────────────────────────────────────────
variable missing complete n mean sd p0 p25 p50 p75 p100
lifeExp 0 142 142 67.01 12.07 39.61 57.16 71.94 76.41 82.6The skim() output now reports summaries for categorical variables (Variable type:factor) separately from the numerical variables (Variable type:numeric). For the categorical variable continent it now reports:
missing,complete,nwhich are the number of missing, complete, and total number of values as before.n_unique: The number of unique levels to this variable, corresponding to Africa, Asia, Americas, Europe, and Oceania.top_counts: In this case the top four counts:Africahas 52 corresponding to its 52 countries,Asiahas 33,Europehas 30, andAmericashas 25. Not displayed isOceaniawith 2 countries.ordered: This tells is whether the categorical variable is “ordinal”: whether there is encoded hierarchy (like low, medium, high). In this case, it is not ordered.
Turning our attention to the summary statistics of the numerical variable lifeExp, we observe that the global median life expectancy is 71.94, or in other words half of the world’s countries (71 countries) will have a life expectancy less than 71.94. The mean life expectancy of 67.01 is lower however. Why is the mean life expectancy lower than the median?
We can answer this question via the last of the three common steps in an exploratory data analysis: creating data visualizations. Let’s visualize the distribution of our outcome variable \(y\) = lifeExp in Figure 6.7.
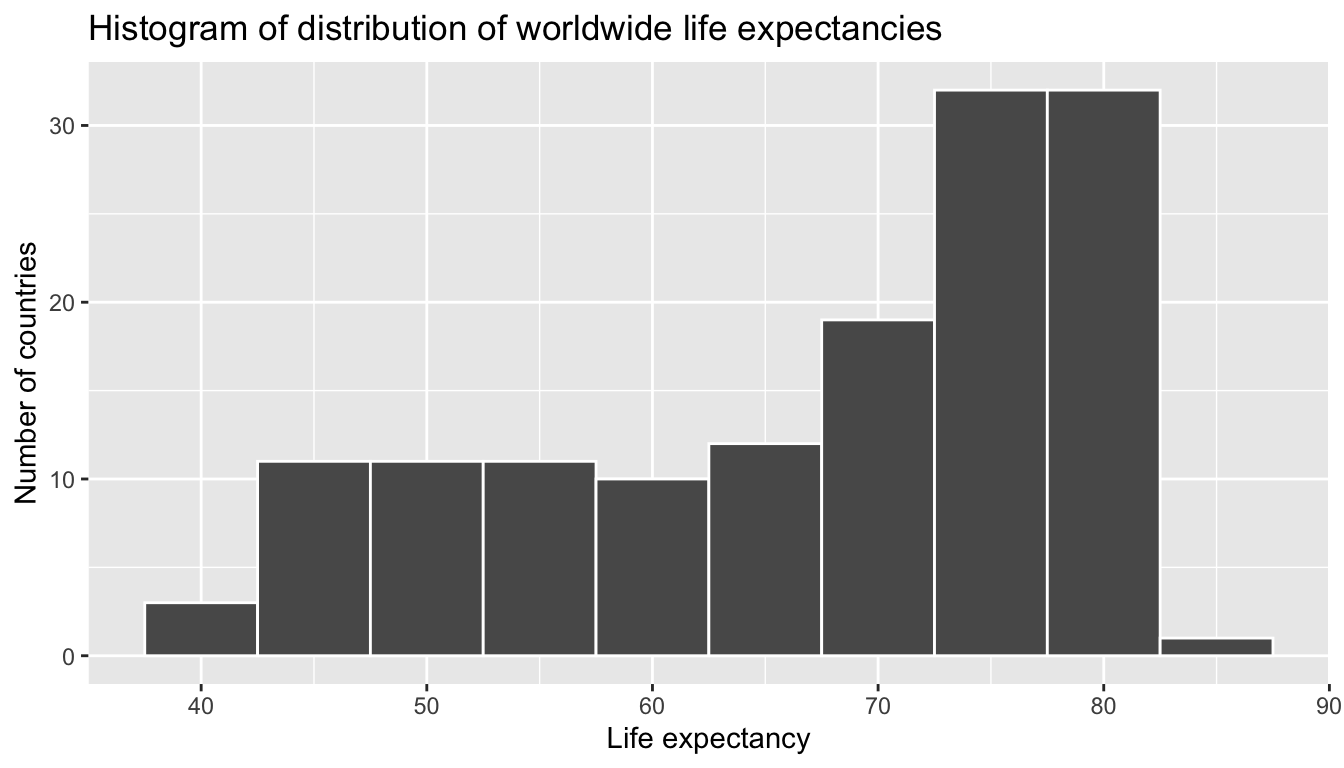
FIGURE 6.7: Histogram of Life Expectancy in 2007.
We see that this data is left-skewed, also known as negatively skewed: there are a few countries with very low life expectancy that are bringing down the mean life expectancy. However, the median is less sensitive to the effects of such outliers, hence the median is greater than the mean in this case.
Remember however, that we want to compare life expectancies both between continents and within continents. In other words, our visualizations need to incorporate some notion of the variable continent. We can do this easily with a faceted histogram. Recall from Section 3.6 that facets allow us to split a visualization by the different values of another variable. We display the resulting visualization in Figure 6.8 by adding a facet_wrap(~ continent, nrow = 2) layer.
ggplot(gapminder2007, aes(x = lifeExp)) +
geom_histogram(binwidth = 5, color = "white") +
labs(x = "Life expectancy", y = "Number of countries",
title = "Histogram of distribution of worldwide life expectancies") +
facet_wrap(~ continent, nrow = 2)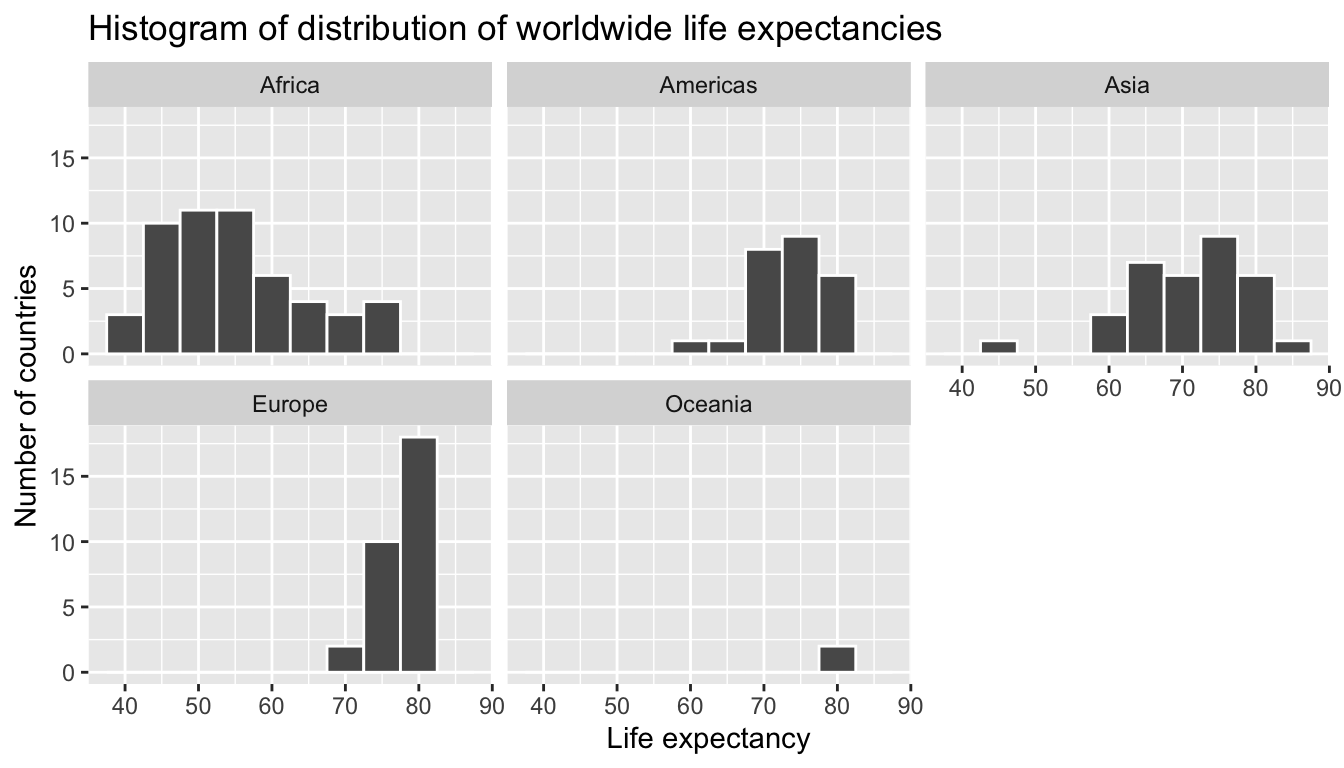
FIGURE 6.8: Life expectancy in 2007.
We observe that unfortunately the distribution of African life expectancies is much lower than the other continents while in Europe life expectancies tend to be higher and do not vary as much. Both Asia and Africa have the most variation in life expectancies. There is the least variation in Oceania, but this greatly influenced by the fact that there are only two countries in Oceania: Australia and New Zealand.
Recall than an alternative visualization of the distribution of a numerical variable split by a categorical variable is by using a side-by-side boxplot via a geom_boxplot(); we map the categorical variable continent to the \(x\)-axis and the different life expectancies within each continent on the \(y\)-axis.
ggplot(gapminder2007, aes(x = continent, y = lifeExp)) +
geom_boxplot() +
labs(x = "Continent", y = "Life expectancy (years)",
title = "Life expectancy by continent")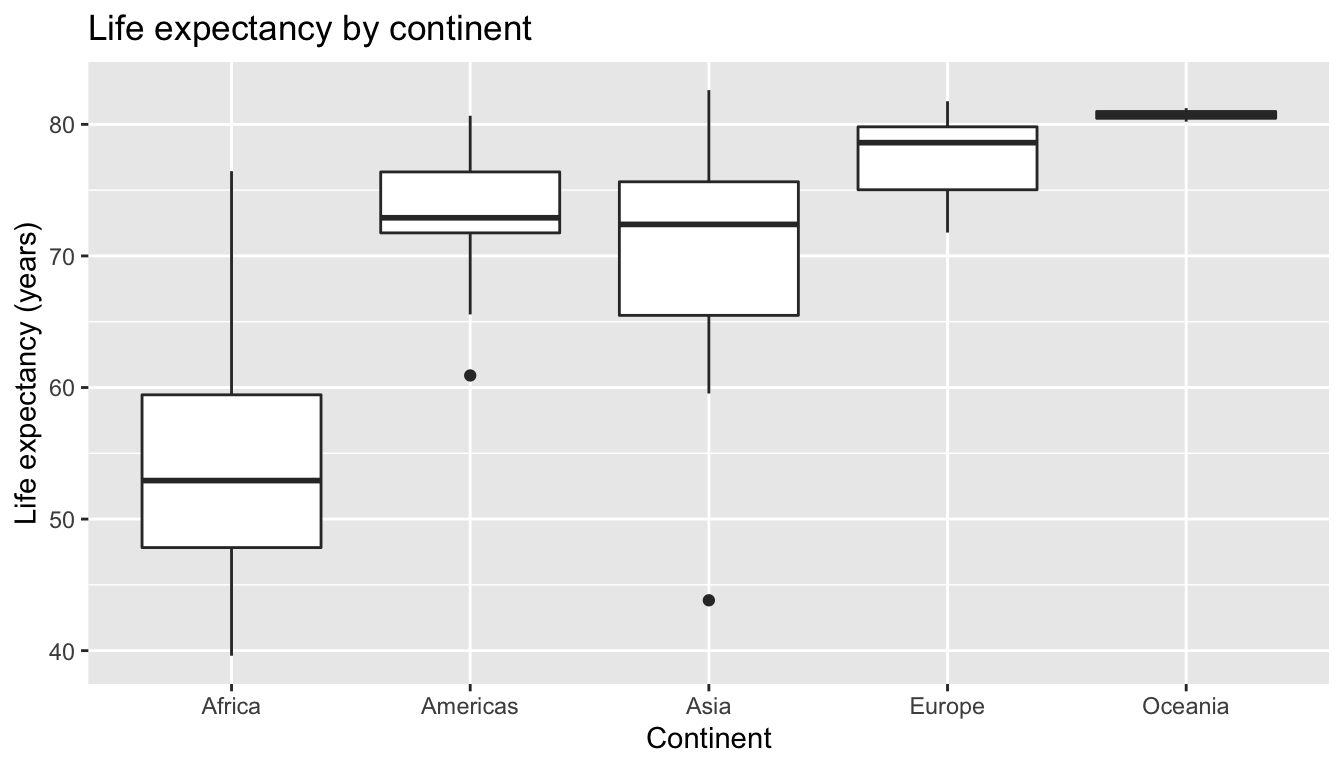
FIGURE 6.9: Life expectancy in 2007.
Some people prefer comparing the distributions of a numerical variable between different levels of a categorical variable using a boxplot instead of a faceted histogram as we can make quick comparisons with imaginary horizontal lines. For example, observe in Figure 6.9 that Oceania clearly tends to have the highest life expectancies given that we could draw an imaginary horizontal line at \(y\) = 80. Furthermore, as we observed with in faceted histogram in Figure 6.8, Africa and Asia have the biggest variation in life expectancy as evidenced by their large interquartile ranges i.e. the height of the boxes.
It’s important to remember however that the solid lines in the middle of the boxes correspond to the medians (i.e. the middle value) rather than the mean (the average). So, for example, if you look at Asia, the solid line denotes the median life expectancy of around 72 years. This indicates to us that half of all countries in Asia have a life expectancy below 72 years whereas half of all countries in Asia have a life expectancy above 72 years.
Let’s compute the median and mean life expectancy for each continent with a little more data wrangling and display the results in Table 6.6.
lifeExp_by_continent <- gapminder2007 %>%
group_by(continent) %>%
summarize(median = median(lifeExp), mean = mean(lifeExp))| continent | median | mean |
|---|---|---|
| Africa | 52.9 | 54.8 |
| Americas | 72.9 | 73.6 |
| Asia | 72.4 | 70.7 |
| Europe | 78.6 | 77.6 |
| Oceania | 80.7 | 80.7 |
Observe the order of the second column median life expectancy: Africa is lowest, the Americas and Asia are next with similar medians, then Europe, then Asia. This ordering corresponds to the ordering of the solid black lines inside the boxes in our side-by-side boxplot in Figure 6.9. Let’s now turn our attention to the values in the third column mean. Using Africa as a baseline for comparison, let’s start making relative comparisons of life expectancies for the other continents:
- The mean life expectancy of the Americas is 73.6 - 54.8 = 18.8 years higher.
- The mean life expectancy of Asia is 70.7 - 54.8 = 15.9 years higher.
- The mean life expectancy of Europe is 77.6 - 54.8 = 22.8 years higher.
- The mean life expectancy of Oceania is 80.7 - 54.8 = 25.9 years higher.
Let’s put these values Table 6.7, which we’ll revisit later on in this section.
| continent | mean | Difference relative to Africa |
|---|---|---|
| Africa | 54.8 | 0.0 |
| Americas | 73.6 | 18.8 |
| Asia | 70.7 | 15.9 |
| Europe | 77.6 | 22.8 |
| Oceania | 80.7 | 25.9 |
Learning check
(LC6.4) Conduct a new exploratory data analysis with the same explanatory variable \(x\) being continent but with gdpPercap as the new outcome variable \(y\). Remember, this involves three things:
- Most crucially: Looking at the raw data values.
- Computing summary statistics, like means, medians, and interquartile ranges.
- Creating data visualizations.
What can you say about the differences in GDP per capita between continents based on this exploration?
6.2.2 Linear regression
In Subsection 6.1.2 we introduced simple linear regression which involves modeling the relationship between a numerical outcome variable \(y\) and a numerical explanatory variable \(x\). In our life expectancy example, we now instead have a categorical explanatory variable \(x\) continent. However our model will not yield a “best-fitting” regression line like in Figure 6.4, but rather “offsets relative to a baseline for comparison.”
As we did in Section 6.1.2 when studying the relationship between teaching scores and beauty scores, let’s output the regression table for this model. Recall that this is done in two steps:
- We first “fit” the linear regression model using the
lm(y~x, data)function and save it inlifeExp_model. - We get the regression table by applying the
get_regression_table()from themoderndivepackage tolifeExp_model.
# Fit regression model:
lifeExp_model <- lm(lifeExp ~ continent, data = gapminder2007)
# Get regression table:
get_regression_table(lifeExp_model)| term | estimate | std_error | statistic | p_value | lower_ci | upper_ci |
|---|---|---|---|---|---|---|
| intercept | 54.8 | 1.02 | 53.45 | 0 | 52.8 | 56.8 |
| continentAmericas | 18.8 | 1.80 | 10.45 | 0 | 15.2 | 22.4 |
| continentAsia | 15.9 | 1.65 | 9.68 | 0 | 12.7 | 19.2 |
| continentEurope | 22.8 | 1.70 | 13.47 | 0 | 19.5 | 26.2 |
| continentOceania | 25.9 | 5.33 | 4.86 | 0 | 15.4 | 36.5 |
Let’s once again focus on the values in the term and estimate columns of Table 6.8. Why are there now 5 rows? Let’s break them down one-by-one:
intercepthere corresponds to the mean life expectancy of countries in Africa of 54.8 years.continentAmericascorresponds to countries in thecontinentof the Americas and the value +18.8 is the same difference in mean life expectancy relative to Africa we displayed in Table 6.7. In other words, the mean life expectancy of countries in the Americas is 54.8 + 18.8 = 73.6.continentAsiacorresponds to countries in thecontinentof Asia and the value +15.9 is the same difference in mean life expectancy relative to Africa we displayed in Table 6.7. In other words, the mean life expectancy of countries in Asia is 54.8 + 15.9 = 70.7.continentEuropecorresponds to countries in thecontinentof Europe and the value +22.8 is the same difference in mean life expectancy relative to Africa we displayed in Table 6.7. In other words, the mean life expectancy of countries in Europe is 54.8 + 22.8 = 77.6.continentOceaniacorresponds to countries in thecontinentof Oceania and the value +25.9 is the same difference in mean life expectancy relative to Africa we displayed in Table 6.7. In other words, the mean life expectancy of countries in the Oceania is 54.8 + 25.9 = 80.7.
In other words, the 5 values in the estimate correspond to:
- The “baseline for comparison” continent Africa (the intercept).
- Four “offsets” from this baseline for the remaining 4 continents: the Americas, Asia, Europe, and Oceania.
You might be asking at this point why was Africa chosen as the “baseline for comparison” group. For no other reason than it comes first alphabetically of the five continents; by default R arranges factors/categorical variables in alphanumeric order. However you can change this baseline group to be another continent if you manipulate the variable continent’s factor “levels” using the forcats package. See Chapter 15 of Garrett Grolemund and Hadley Wickham’s book (Grolemund and Wickham 2016) for examples.
Let’s now write the equation for our fitted values \(\widehat{y} = \widehat{\text{life exp}}\).
\[ \begin{aligned} \widehat{y} = \widehat{\text{life exp}} &= b_0 + b_{\text{Amer}}\cdot\mathbb{1}_{\mbox{Amer}}(x) + b_{\text{Asia}}\cdot\mathbb{1}_{\mbox{Asia}}(x) + \\ & \qquad b_{\text{Euro}}\cdot\mathbb{1}_{\mbox{Euro}}(x) + b_{\text{Ocean}}\cdot\mathbb{1}_{\mbox{Ocean}}(x)\\ &= 54.8 + 18.8\cdot\mathbb{1}_{\mbox{Amer}}(x) + 15.9\cdot\mathbb{1}_{\mbox{Asia}}(x) + \\ & \qquad 22.8\cdot\mathbb{1}_{\mbox{Euro}}(x) + 25.9\cdot\mathbb{1}_{\mbox{Ocean}}(x) \end{aligned} \]
Whoa! That looks very daunting! Don’t fret however, as once you understand what all the elements mean, things simply greatly. First, \(\mathbb{1}_{A}(x)\) is what’s known in mathematics as an “indicator function” that takes only one of two possible values, 0 and 1, where
\[ \mathbb{1}_{A}(x) = \left\{ \begin{array}{ll} 1 & \text{if } x \text{ is in } A \\ 0 & \text{if } \text{otherwise} \end{array} \right. \]
In a statistical modeling context this is also known as a “dummy variable”. In our case, let’s consider the first such indicator variable; this indicator function returns 1 if a country is in the Americas, 0 otherwise.
\[ \mathbb{1}_{\mbox{Amer}}(x) = \left\{ \begin{array}{ll} 1 & \text{if } \text{country } x \text{ is in the Americas} \\ 0 & \text{otherwise}\end{array} \right. \]
Second, \(b_0\) corresponds to the intercept as before; in this case its the mean life expectancy of all countries in Africa. Third, the \(b_{\text{Amer}}\), \(b_{\text{Asia}}\), \(b_{\text{Euro}}\), and \(b_{\text{Ocean}}\) represent the 4 “offsets relative to the baseline for comparison” in the regression table output in Table 6.8: continentAmericas, continentAsia, continentEurope, and continentOceania.
Let’s put this all together and compute the fitted value \(\widehat{y} = \widehat{\text{life exp}}\) for a country in Africa. Since the country is in Africa, all four indicator functions \(\mathbb{1}_{\mbox{Amer}}(x)\), \(\mathbb{1}_{\mbox{Asia}}(x)\), \(\mathbb{1}_{\mbox{Euro}}(x)\), and \(\mathbb{1}_{\mbox{Ocean}}(x)\) will equal 0, and thus:
\[ \begin{aligned} \widehat{\text{life exp}} &= b_0 + b_{\text{Amer}}\cdot\mathbb{1}_{\mbox{Amer}}(x) + b_{\text{Asia}}\cdot\mathbb{1}_{\mbox{Asia}}(x) + \\ & \qquad b_{\text{Euro}}\cdot\mathbb{1}_{\text{Euro}}(x) + b_{\text{Ocean}}\cdot\mathbb{1}_{\text{Ocean}}(x)\\ &= 54.8 + 18.8\cdot\mathbb{1}_{\text{Amer}}(x) + 15.9\cdot\mathbb{1}_{\text{Asia}}(x) + \\ & \qquad 22.8\cdot\mathbb{1}_{\text{Euro}}(x) + 25.9\cdot\mathbb{1}_{\text{Ocean}}(x)\\ &= 54.8 + 18.8\cdot 0 + 15.9\cdot 0 + 22.8\cdot 0 + 25.9\cdot 0\\ &= 54.8 \end{aligned} \]
In other words, all that’s left is the intercept \(b_0\) corresponding to the average life expectancy of African countries of 54.8 years. Next, say we are considering a country in the Americas. In this case only the indicator function \(\mathbb{1}_{\mbox{Amer}}(x)\) for the Americas will equal 1, while all the others will equal 0, and thus:
\[ \begin{aligned} \widehat{\text{life exp}} &= 54.8 + 18.8\cdot\mathbb{1}_{\mbox{Amer}}(x) + 15.9\cdot\mathbb{1}_{\mbox{Asia}}(x) + 22.8\cdot\mathbb{1}_{\mbox{Euro}}(x) + \\ & \qquad 25.9\cdot\mathbb{1}_{\mbox{Ocean}}(x)\\ &= 54.8 + 18.8\cdot 1 + 15.9\cdot 0 + 22.8\cdot 0 + 25.9\cdot 0\\ &= 54.8 + 18.8\\ &= 72.9 \end{aligned} \]
which is the mean life expectancy for countries in the Americas of 72.9 years we computed in the previous subsection in Table 6.7. Note the “offset from the baseline for comparison” here is +18.8 years. Let’s do one more. Say we are considering a country in Asia. In this case only the indicator function \(\mathbb{1}_{\mbox{Asia}}(x)\) for Asia will equal 1, while all the others will equal 0, and thus:
\[ \begin{aligned} \widehat{\text{life exp}} &= 54.8 + 18.8\cdot\mathbb{1}_{\mbox{Amer}}(x) + 15.9\cdot\mathbb{1}_{\mbox{Asia}}(x) + 22.8\cdot\mathbb{1}_{\mbox{Euro}}(x) + \\ & \qquad 25.9\cdot\mathbb{1}_{\mbox{Ocean}}(x)\\ &= 54.8 + 18.8\cdot 0 + 15.9\cdot 1 + 22.8\cdot 0 + 25.9\cdot 0\\ &= 54.8 + 15.9\\ &= 70.7 \end{aligned} \]
which is the mean life expectancy for countries in the Americas of 72.9 years we computed in the previous subsection in Table 6.7. Note the “offset from the baseline for comparison” here is +15.9 years.
Let’s generalize this idea a bit. If we fit a linear regression model using a categorical explanatory variable \(x\) that has \(k\) levels i.e. possible categories, a regression model will return an intercept and \(k - 1\) “offsets.” In our case, since there are \(k = 5\) continents, the regression model returns an intercept corresponding to the baseline for comparison Africa and \(k - 1 = 4\) offsets corresponding to the Americas, Asia, Europe, and Oceania.
Phew! That was a lot of work! Understanding a regression table output when you’re using a categorical explanatory variable is a topic that many new regression practitioners struggle with. The only real remedy for these struggles is practice, practice, practice. However, once you equip yourselves with an understanding of how to create regression models using categorical explanatory variables, you’ll be able to incorporate many new variables in your models, given the large amount of the world’s data that is categorical. If you feel like you’re still struggling at this point however, we suggest you closely compare Tables 6.7 and 6.8 and note how you can compute all the values from one table using the values in the other.
Learning check
(LC6.5) Fit a new linear regression using lm(gdpPercap ~ continent, data = gapminder2007) where gdpPercap is the new outcome variable \(y\). Get information about the “best-fitting” line from the regression table by applying the get_regression_table() function. How do the regression results match up with the results from your previous exploratory data analysis?
6.2.3 Observed/fitted values and residuals
Recall in Subsection 6.1.3, we defined the following three concepts:
- Observed values \(y\), or the observed value of the outcome variable
- Fitted values \(\widehat{y}\), or the value on the regression line for a given \(x\) value
- Residuals \(y - \widehat{y}\), or the error between the observed value and the fitted value
We obtained these values and other values using the get_regression_points() function from the moderndive package. This time however, let’s add an ID = "country" argument: this is telling the function to use the variable country in gapminder2007 as an identification variable in the output. This will help contextualize our analysis by matching values to countries.
regression_points <- get_regression_points(lifeExp_model, ID = "country")
regression_points| country | lifeExp | continent | lifeExp_hat | residual |
|---|---|---|---|---|
| Afghanistan | 43.8 | Asia | 70.7 | -26.900 |
| Albania | 76.4 | Europe | 77.6 | -1.226 |
| Algeria | 72.3 | Africa | 54.8 | 17.495 |
| Angola | 42.7 | Africa | 54.8 | -12.075 |
| Argentina | 75.3 | Americas | 73.6 | 1.712 |
| Australia | 81.2 | Oceania | 80.7 | 0.515 |
| Austria | 79.8 | Europe | 77.6 | 2.180 |
| Bahrain | 75.6 | Asia | 70.7 | 4.907 |
| Bangladesh | 64.1 | Asia | 70.7 | -6.666 |
| Belgium | 79.4 | Europe | 77.6 | 1.792 |
Observe in Table 6.9
lifeExp_hatare the fitted values \(\widehat{y}\) = \(\widehat{\text{lifeexp}}\).- If you look closely, there are only 5 possible values for
lifeExp_hat. These correspond to the 5 mean life expectancies for the 5 continents we displayed in Table 6.7: Africa 54.8, the Americas 73.6, Asia 70.7, Europe 77.6, and Oceania 80.7 - The
residualcolumn is simply \(y - \widehat{y}\) =lifeexp - lifeexp_hat. These values can be interpreted the deviation of a country’s life expectancy from it’s continent’s average life expectancy. For example, look at the the first row Table 6.9 corresponding to Afghanistan. The residual of \(y - \widehat{y}\) = 43.8 - 70.7 = -26.9 is indicating that Afghanistan’s life expectancy is a whopping 26.9 years lower than the mean life expectancy of all Asia countries. This can in part be explained by the many years of war that country has suffered.
Learning check
(LC6.6) Using either the sorting functionality of RStudio’s spreadsheet viewer or using the data wrangling tools you learned in Chapter 4, identify the 5 countries with the 5 smallest (most negative) residuals? What do these negative residuals say about their life expectancy relative to their continents?
(LC6.7) Repeat the above, but identify the 5 countries with the 5 largest (most positive) residuals. What do these negative residuals say about their life expectancy relative to their continents?
6.4 Conclusion
6.4.1 Additional resources
An R script file of all R code used in this chapter is available here.
As we suggested in Subsection 6.1.1, interpreting coefficients that are not close to the extreme values of -1 and 1 can be subjective. To develop your sense of correlation coefficients, we suggest you play the following 80’s-style video game called “Guess the correlation” at http://guessthecorrelation.com/.
FIGURE 6.14: Preview of “Guess the Correlation” Game.
6.4.2 What’s to come?
In this chapter, you’ve studied what we like to term “basic regression” where you only have one explanatory variable. In Chapter 7, we’ll study multiple regression where our regression models can have more than one explanatory variable! In particular, we’ll consider two scenarios: regression models with one numerical and one categorical explanatory variables and regression models with two numerical explanatory variables. This will allow you to construct more sophisticated and powerful models in the hopes of better explaining your outcome variable \(y\) of interest.
References
Grolemund, Garrett, and Hadley Wickham. 2016. R for Data Science. http://r4ds.had.co.nz/.